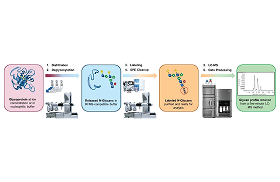
- Amino Acid Analysis
- Bioprocessing
- Cell and Gene Therapy
- DMPK: Automated Sample Preparation
- ELISA Assays
- Endotoxin Testing
- Forensic Toxicology
- Genomics
- Immunosuppressant Drug Monitoring
- Lipidomics
- Metabolomics
- Oligonucleotide Therapies
- Pesticide Analysis
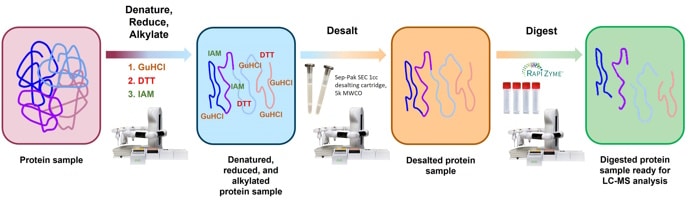
- Protein Therapies
- Proteomics
- RNA Therapeutics
- RT-qPCR Testing
- Small Molecule Drugs
- Amino Acid Analysis
- Bioprocessing
- Cell and Gene Therapy
- DMPK: Automated Sample Preparation
- ELISA Assays
- Endotoxin Testing
- Forensic Toxicology
- Genomics
- Immunosuppressant Drug Monitoring
- Lipidomics
- Metabolomics
- Oligonucleotide Therapies
- Pesticide Analysis
- Protein Therapies
- Proteomics
- RNA Therapeutics
- RT-qPCR Testing
- Small Molecule Drugs
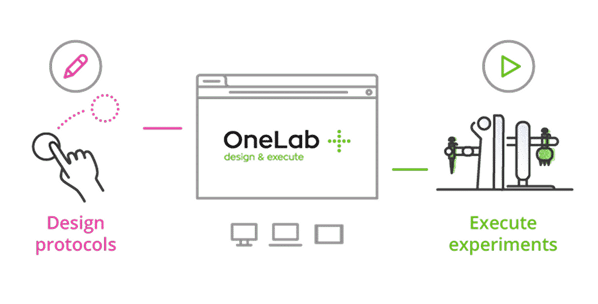
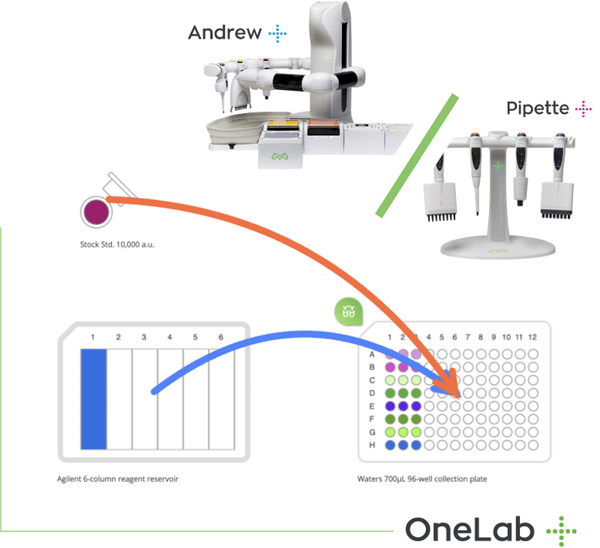
Master Serial Dilutions with Ease
From column-wise to row-wise preparation, streamline your dilution workflows with OneLab and Andrew+ for unmatched precision and reproducibility.
Looking for product support or a quote? We’re here to help. Contact Us