Simple and Efficient Aliquotting
Andrew is the most versatile and economically effective robotic platform to automate the most ordinary lab activity – aliquotting. Besides reducing hands-on time for repetitive aliquotting procedures by one order of magnitude, the Andrew robot and the user-friendly software Andrew Lab efficiently accommodate any aliquotting strategy with your existing consumables, all this while assuring perfect traceability and avoiding mistakes.
The aliquotting scenarios
The term “aliquot” has different meanings in different fields: in biology and chemistry, it refers to a smaller fraction of a whole sample. In most cases, only these fractions are needed for an experiment, hence aliquotting must be done before other productive tasks can be delivered. Aliquotting can be just the simple transfer of the same volume from one source to multiple destinations, but it can also involve a more complex distribution from multiple sources to multiple destinations, at different volumes and different concentrations. Further complexity can occur when replicates for each different aliquot are required. As the task becomes more complex, so does the required amount of hands-on time from lab personnel and the opportunity for protocol execution errors and variability to occur. The repetitive aliquotting task also exposes users to the occurrence of Repetitive Stress Injuries (RSI), and finally all these aspects have a significant impact on the cost and quality of laboratory activities. Many traditional automated liquid handling systems are available today. However, their systematic use is often neither cost-effective nor accessible for aliquotting activities: apart from the time-consuming setup of an expensive and complex device, most automated liquid handlers require specific and particularly costly labware designed for automation an unjustified extra cost if the purpose is aliquotting samples to be used in manual experiments.
Perhaps the most common application of aliquotting is to extract the original stock samples into smaller portions for different analytical tests of the same materials while preserving the original stock for future usage. This takes place daily at biobanks where thousands of body fluid samples such as blood, urine, and saliva are delivered in tubes that must
be aliquoted into multiple cryo-vials. Another common purpose is to aliquot master mixes of reagents and buffers during sample preparation for biochemical and genetic tests in molecular diagnostic, forensic, and pre-clinical assays, such as ELISA, PCR, and RT-qPCR. In these cases, the aliquots are often taken from individual tubes into an array of smaller tubes, 96- or 384-well microplates. Aliquotting is also used to avoid repeated freeze-thaw or open-close cycle of reagents which helps prevent changes in the physicochemical characteristics, such as easily oxidized chemistries of dNTPs, heat-labile enzymes, light-sensitive fluorescence conjugating moieties, volatile solvents, and delicate living cells. Only small aliquots of these reagents are needed at a time, and using individual aliquots only when required minimizes their quality degradation and the consequence of failed experiments and costly repetition of work.
Although many aliquotting applications require just a single volume to be dispensed over and over, some require continuous changing of volumes. This is the case, for example, of lyophilized primer samples, which are normally synthesized at different amounts in microtubes and thus must be dissolved at different volumes to equalize the concentration of all primer stock solutions. Likewise, normalization of different DNA, RNA or protein samples to the same concentration is a common task that requires aliquotting different volumes of water or buffer to the original sources or to different destinations. Aliquotting variable volumes is particularly error-prone, time consuming, and tedious for laboratory personnel to carry out. Human variability and simple errors in aliquotting can be highly detrimental, leading to unusable experimental results and unrepairable waste of valuable samples or reagents.
Manual aliquotting completely eliminated by Andrew
Automating the aliquotting process is the most obvious and convenient choice to solve the problems of decreased lab efficiency, too-busy working days, pipetting mistakes, and increased occurrence of RSI. However, most automated systems available to date are compatible with only limited consumables specifically designed for such system, ignoring the multiplicity of source and destination labware actually used in laboratories and consequently requiring additional transfer steps and reformatting of samples. Thanks to its highly adaptable working deck, the bench-top pipetting robot Andrew is the most affordable and suitable solution for small to medium throughput pipetting activities, offering the consumable flexibility not encountered with other traditional robotic stations. Andrew’s working deck consists of an innovative system of magnetic tiles called Domino blocks, which can be adapted to virtually any type of regular consumable used in aliquotting tasks. For this reason, Andrew can aliquot in nearly any arrangement defined by users while also can perform more complex liquid handling tasks such as serial dilutions, cross-reactions, normalization, array reformatting, creation of master plates, and cherry picking. Operated from the user-friendly software Andrew Lab, Andrew uses the same single channel pipettes present in almost all life-science laboratories. Aliquotting protocols can be set up within minutes with Andrew Lab, minimalizing the time spent during protocol design. Additional error-free traceability and documentation is completed by importing and exporting data from and to LIMS, ELNs, or any other spreadsheet format.
How to aliquot with Andrew? Three examples
1. Aliquotting from one source tube to multiple destination tubes with the same volume
Thanks to the “Repetitive” mode of Andrew, one single aspirated volume of a solution can be rapidly dispensed in multiple destination tubes. Using standard, classical mechanical pipettes, Andrew fills up the pipette tip to the highest capacity and dispenses multiple aliquots of the same volume in succession without touching the liquid. Therefore, it can go back to the source without contamination risk, to refill for subsequent multi-dispensing cycles. To ensure both accuracy and precision, Andrew will discard the first and last aliquots of the dispensed series, which normally are affected by the largest error. For example, three 200 μL-aliquots of a given stock sample can be created with only one aspiration step using a 1000 μL pipette (Figure 1). By applying the same principle to 18 aliquots of 50 μL done with a single aspiration step, the advantage of repetitive mode becomes obvious: it significantly shortens the time needed to aliquot compared to single dispensing and minimizes the consumption of tips (see the Andrew Alliance Application Note on Repetitive dispensing in Reference).
The Andrew productivity in aliquotting from one source to multiple destination tubes can be exemplified by two common procedures performed in many diagnostic labs: preparing aliquots of whole blood and urine samples (Figure 2). In the virtual lab bench space of Andrew Lab, the user simply indicates the pipetting direction from the source tube “blood” to the first destination tube “cryo” with the mouse cursor, and holding down the Ctrl key while tapping additional tubes will include additional aliquotting targets. Then, it is enough to enter the aliquotting volume in the pop-up screen. The same process can be repeated for aliquotting from the source tube “urine” to the 4 destination tubes “vial”, by taking less than 2 minutes in Andrew Lab. Moreover, Andrew Lab protocols can be saved and re-used for recurrent aliquotting tasks, which obviously improves efficiency when handling a large number of destinations.

Figure 1: Schematic of the Repetitive pipetting mode of Andrew, Figure 2: Example of an Andrew Lab protocol for aliquotting blood and urine samples with Andrew
2. Aliquotting from one source tube to multiple destination tubes with different volumes
Another very common situation in the lab is the aliquotting of variable volumes from one source to multiple destination tubes. An example of this scenario is the preparation of primer stocks. Since synthesis yields vary between oligonucleotide sequences and experiments, researchers must dissolve the lyophilized primer stocks with different volumes of water or buffer to have them all at the same concentration. Using Andrew Lab and Andrew, the experimental design and execution of such task become extremely simple and error-free: the process starts by the creation of the desired consumables corresponding to the source (water or buffer) and destination vials in the virtual bench space of Andrew Lab (Figure 3A), then is followed by the addition of a pipetting step. This can be accomplished by simply dragging the mouse cursor from the source tube to one of the destination tubes or by clicking on the “Pipetting” action from the “Add step” panel. Next, multiple volumes can be indicated by entering the volumes of buffer to be pipetted to each primer tube in the list that will be displayed after activating the “Multi-volume mode” option (Figure 3B). For time optimization purposes, the volumes can be entered in ascendant or descendent order, as this will minimize the time required for Andrew to change volumes and tips. Please remark that the paste option allows to quickly introduce the values all at once, by copying and pasting from a list previously designed in a spreadsheet or another document (Table 1). The textual and graphical description of this aliquotting activity will appear as a single instruction (Figure 3C).
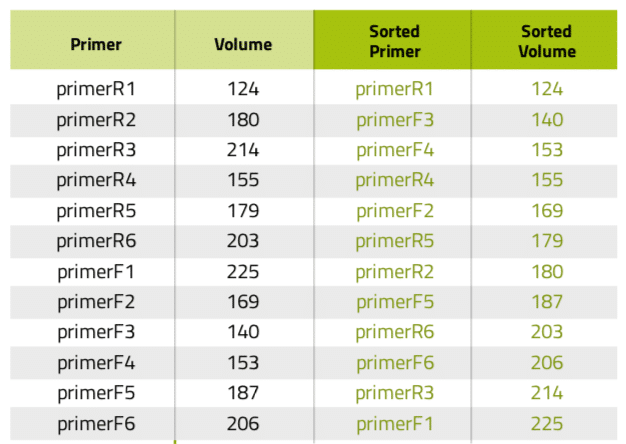
Table 1: Primer list before and after being sorted according to volume.
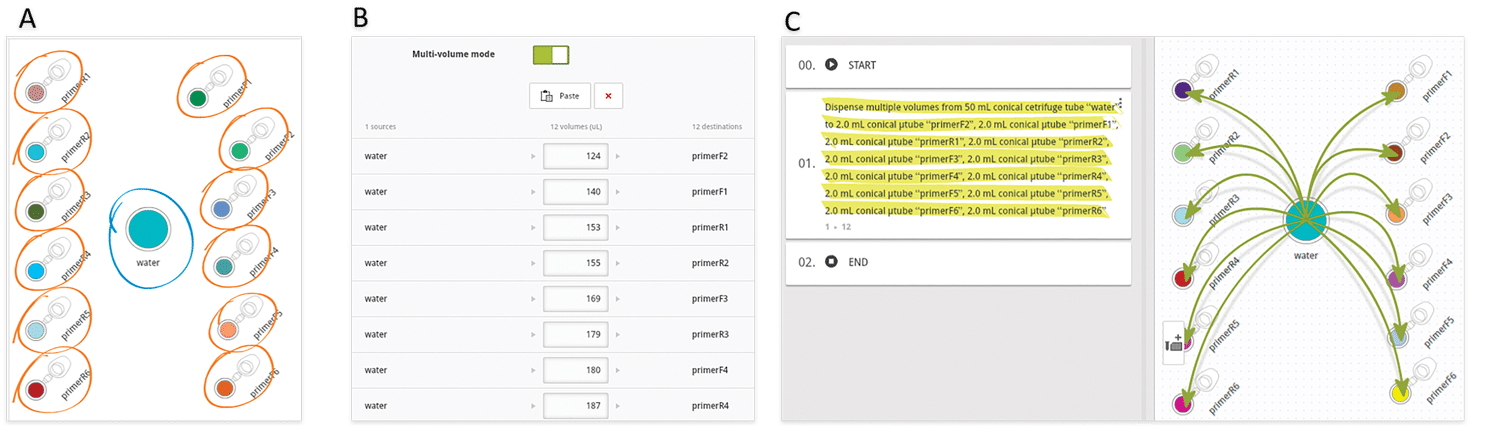
Figure 3: Main steps in designing an aliquotting experiment from one source to many destination tubes in Andrew Lab
The subsequent resuspension step can also be automated with Andrew. To do so, users can introduce a mixing action at the destination tubes under the “Mixing” panel. The speed of this mixing is adaptable (slow, normal, or fast) to fit the user preference and/or experiment requirements. Introduction of this mixing step can replace the vortexing and centrifuging steps often done during resuspension of the primers, leading to larger time savings and to more reproducibility. Designing this example aliquot protocol took only 2 minutes and executing it with Andrew takes only 7 minutes (with the “on-the-fly” mode of pipetting activated). If the total aliquot number is 100, the hands-on time increases to 10 minutes for designing the protocol plus 5 minutes to set up the consumables for Andrew. On the other hand, manual aliquotting of 100 different volumes requires approximately 50 minutes of hands- on time to finish. Thus, even if tubes must be manually uncapped and accommodated in the Andrew physical work bench, primers stock preparation using Andrew is clearly advantageous as it decreases hands-on time for repetitive aliquotting (using an already designed experimental protocol) by a factor of 10 (from 50 minutes to 5 minutes, Figure 4) and allows a higher number of samples to be processed in a uniform and reproducible fashion.
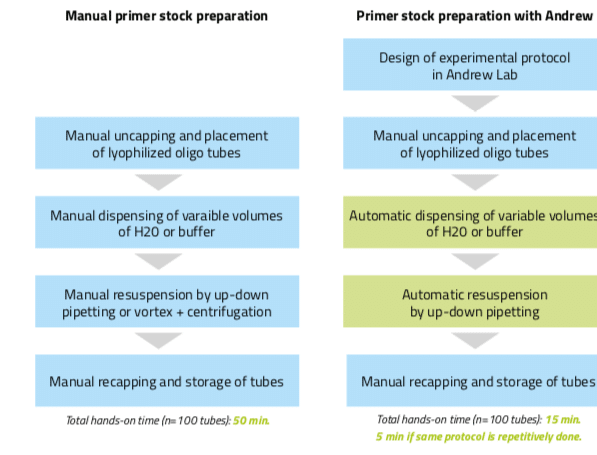
Figure 4: Workflow for the preparation of primer stocks either manually or by using the pipetting robot Andrew
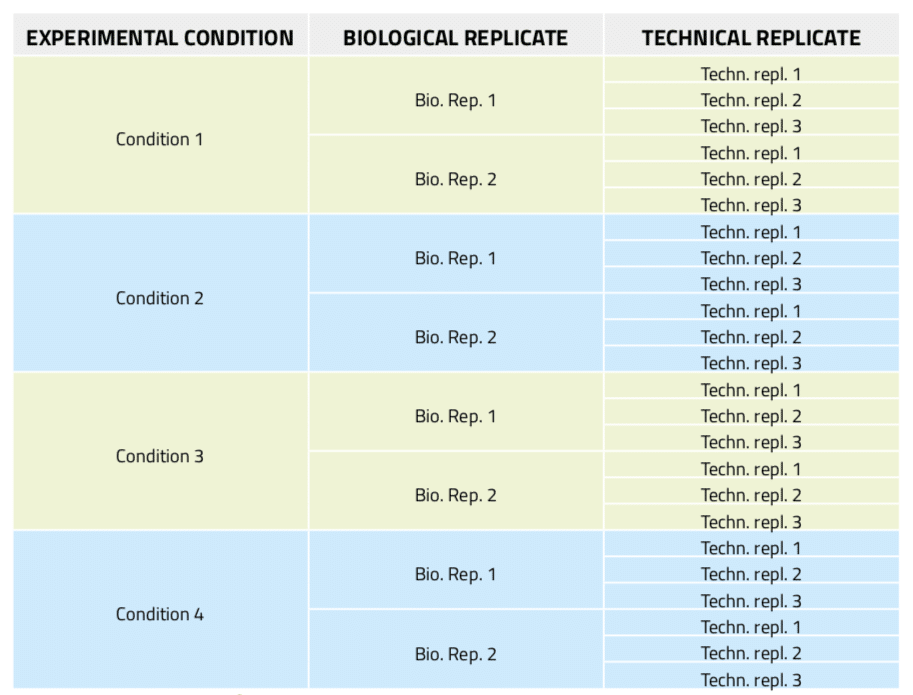
Figure 5: Experimental design of a real-time PCR experiment including three technical replicates per sample.
3. Aliquot from many source tubes to many destinations with replicates
In real-time PCR experiments, technical replicates in addition to biological repeats of the samples are essential to establish the experimental error due to the amplification and analysis technique, thus allowing the setting of confidence levels for what is meant to be significant data. Nonetheless, the experiment preparation becomes more complex and cumbersome as it involves extremely careful and time-consuming aliquotting activities. Thus, automation of this task is much desired. In this example, 8 different DNA samples from 4 different biological conditions with two biological replicates each are tested. For each of these samples, three technical replicates are included, which results in a total of 24 aliquots from variable sources to be prepared (Figure 5). Furthermore, in order to reduce bias error, these reactions must be randomly distributed in a 96-well microplate.
As in the above example, we envisage sorting the volumes to be pipetted to minimize the volume changes, and therefore the experiment execution time. After source and destination tubes have been dropped in triplicates from the virtual bench to the consumables section (Figure 6A), the two lists of the destination well positions and volumes are simply copied and pasted into the corresponding Andrew Lab fields (Figure 6B). All 24 aliquot pipetting steps are consolidated into a single instruction (Figure 6C) and Andrew will change tip for each pipetting step. However, it is possible to use fewer tips (e.g. one single tip for each set of triplicates) in occasions where the user wishes to reduce consumable costs. In this case, the protocol can also be designed as an independent pipetting instruction for each sample, which allows the user to have full control of tip choice for the aliquotting of each sample (Figure 7).
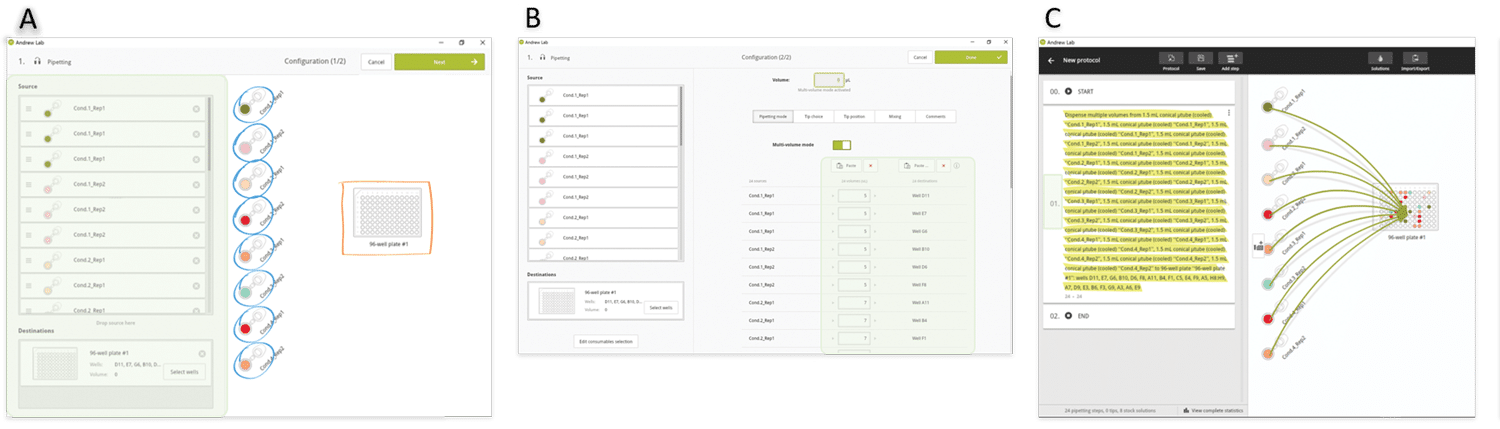
Figure 6: Design of an aliquotting experiment involving many sources and many destination wells in Andrew Lab. A) Source and destination tubes are dropped in the Andrew Lab virtual bench. B) The list of volumes to be pipetted and destination wells positions can be directly introduced or simply pasted from a spreadsheet into the Andrew Lab columns dedicated for this information (green highlighted area). C) At the end of this procedure, all pipetting steps are consolidated in a single instruction (yellow highlighted area).»
If the sources also come from multiple wells in another microplate (or even the same microplate), the entire source plate can be dropped into Source, and the sorted list of source wells positions corresponding to the correct aliquot volumes is then copied and pasted during the Configuration step. The same procedure can be followed if a consumable representing a rack of tubes in any format is dragged onto the virtual bench, making the experimental design easier and faster.
When tips are changed for every single aliquot, this experiment, which takes approximately 5.5 minutes to design in Andrew Lab, is completed fully unattended and without mistakes by Andrew in 10 minutes. While manual execution may take approximately the same time, it is completely operator dependent and prone to possible errors.
Summary
The robot Andrew significantly improves the aliquotting efficiency by minimizing manual labor time, fully eliminating human variations and errors, integrating multiple aliquotting strategies and adapting to a large diversity of labware. Simple-to-design protocols and improved traceability and documentation of the aliquotting steps and materials are facilitated by the intuitive software Andrew Lab. Moreover, it protects the personnel health by avoiding the direct handling of biologically or chemically hazardous liquids, and by reducing the risk of developing muscular disorders such as RSI (see the Andrew Alliance Application Note on pipetting ergonomics in Reference).