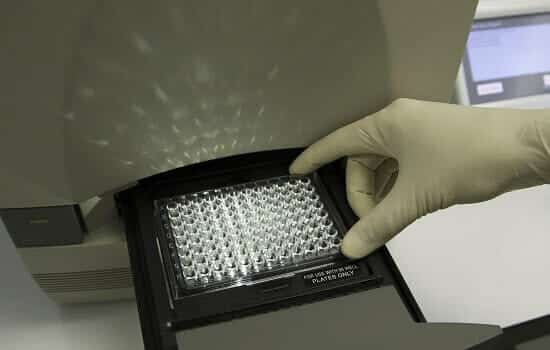
Sample Preparation for Real-Time Quantitative PCR
The pipetting robot Andrew prepares quantitative PCR assays with excellent precision and accuracy. Andrew significantly improves the data quality, operates contamination-free and reduces hands-on preparation. work at the bench from 25 minutes to 2 minutes
Challenges of qPCR sample preparation and automation solution.
Gene expression analysis and diagnostic testing often rely on real-time PCR (qPCR), a powerful technique that quantitates the amount of DNA in the sample [1]. The amplified DNA is detected in real-time by the incorporated fluorescent dye such as SYBR Green or dye-labeled oligo primers such as TaqMan probes. The sensitivity of the dyes at the picogram level renders the detection dynamic range of ≥6 folds. Because PCR products are amplified exponentially, inconsistent pipetting or cross-contamination with a minute amount of DNA can dramatically skew the data and the quantification of DNA, thus invalidating the final results.
For this reason, qPCR experiments require extremely high accuracy and their reproducibility needs to be guaranteed, something particularly challenging when doing them manually, as different pipetting techniques vary among and within operators. These differences are not easily rectified and require correct actions: the pipettes need to be held at vertical position, the pipetting speed should be not too fast especially for viscous solutions of the master mix to avoid splashing the samples, tip submergence should be at the correct level beneath the liquid surface to avoid excess liquid coating outside the tip being transferred inadvertently during dispensing, and the sample needs to be mixed well. Often many replicates are required for statistical inference, demanding the accurate handling of multiple micro-centrifuge tubes and small wells in microplates. These challenging, concentration dependent, and variable pipetting factors often cause mistakes during the tedious qPCR sample preparation.
A robot that handles all pipetting tasks like a human operator without making any mistakes, offering excellent accuracy and reproducibility, will not only eliminate these error sources but also significantly decrease the hands-on time required for scientists and technicians to prepare samples for the qPCR experiments.
Andrew Alliance provides such a pipetting robot, Andrew (Fig. 1), as the most easy-to-use and affordable automating solution for the preparation of qPCR reactions.
Using conventional single channel pipettes designed for manual operations, Andrew is able to deploy the full volume range of its pipettes (from 0.2μl to 1ml) completely unattended [2]. Accompanied by Andrew Lab, the graphical user friendly software that allows the users to design their own pipetting protocol with ease, Andrew is able to smoothly execute all pipetting operations (choosing the right pipette for the volume, setting the volume, inserting and ejecting tips, pre-wetting tips, changing or keeping a tip, mixing by pipetting up and down…) and incubations.
In this application note, we describe the quantification of gene expression in human cells, using Andrew as an automating solution. We specifically measured the expression of 3 genes encoding the human tumor necrosis factor receptor family members and calculated the Threshold Cycle (Ct) which is the inverse proportion of the starting cDNA amounts in the sample, the pipetting accuracy in terms of linear regression from the Ct values of serial dilutions of the cDNA templates, and the pipetting reproducibility quantified in terms of a coefficient of variation (CV) of Ct values from technical replicates. The same experiment with the same materials was executed manually in parallel in order to compare the performance of Andrew with that of a skilled human operator. The results demonstrate that Andrew reduces hands-on working time to more than half while significantly improving data accuracy and reproducibility.
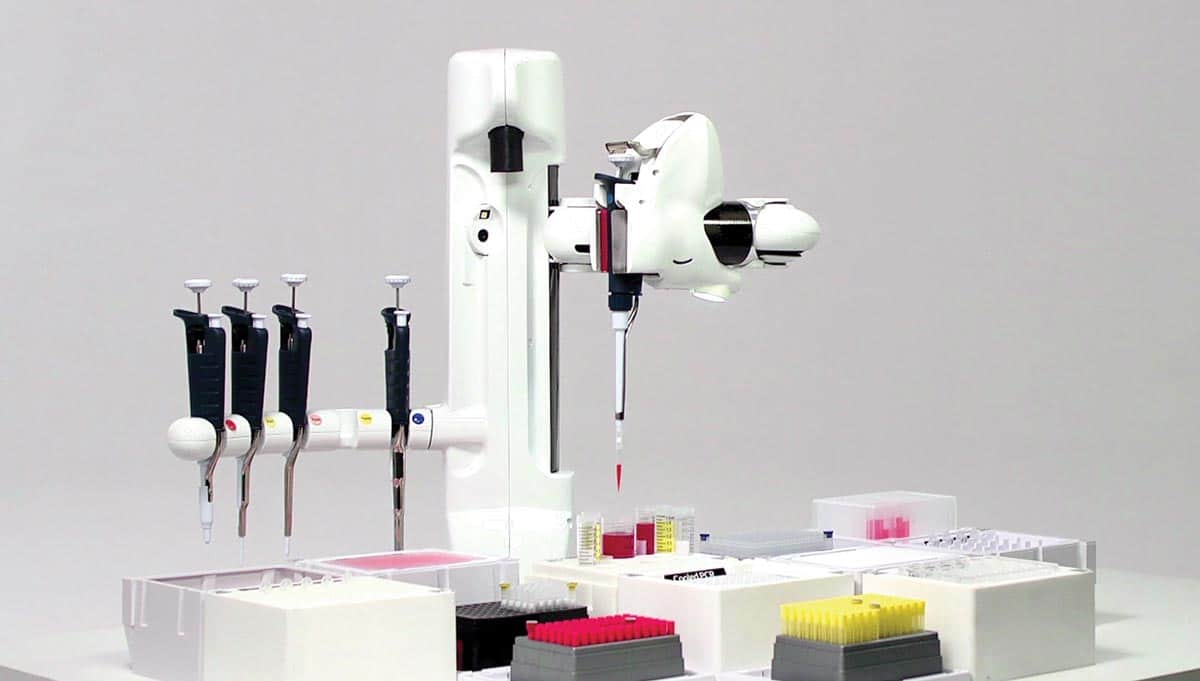
Figure 1: Andrew, the pipetting robot using conventional pipettes
Andrew reduces the hands-on time for qPCR sample preparation

Figure 2: Statistics of pipettes and tips given in Andrew Lab, Figure 3: Top view of working deck of Andrew
Three members of the tumor necrosis factor receptor family were tested on a cDNA extracted from the spleen of C57BL/6J mouse: • TACI (transmembrane activator and calcium-modulator and cyclophilin ligand interactor) • BCMA (B cell maturation) • BAFF Receptor (B-cell activating factor Receptor) ß-actin was used as the positive control, and water as the negative control. The cDNA was diluted at 20, 100, 1000 and 10000 times using the “Serial Dilution” wizard in Andrew Lab. Then 5μl of each dilution were distributed in a 96-well PCR plate (Scientific Specialties Inc, Lodi, CA, USA) using one tip for each dilution. Four different master mixes containing Taq Polymerase, IQ™ SYBR® Green Supermix (Bio-Rad), forward and reverse primers of 4 different primer pairs for the 4 genes were prepared. 13μl of each master mix were added to each point of the cDNA dilution series. Every reagent was mixed when added to the tubes or the wells and the option “high viscosity” was selected in order to avoid bubbles during the preparation and the distribution of the master mix in the PCR plate.
To check the reproducibility of the pipetting operations, all reactions were tested in duplicate or triplicate. Designing the qPCR protocol in the software Andrew Lab took 10 minutes, and the protocol provides a previewed statistics of the types and the numbers of the pipettes and pipette tips needed for users to organize and prepare for the experiments (Fig. 2). Manually placing the consumables onto Andrew’s deck (Fig. 3) took only 2 minutes, and Andrew executed the full protocol unattended without problems. The hands-on work of this protocol with manual operation included 5 minutes of protocol design and 25 minutes of pipetting execution. Thus, using Andrew reduces the total actual bench working time for users from 25 minutes to 2 minutes for this protocol (Fig. 4).
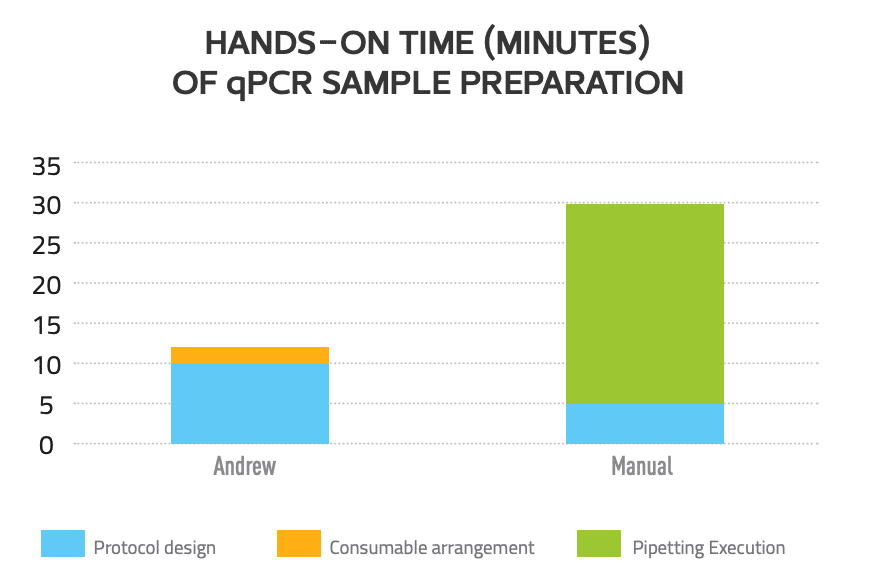
Andrew reduces hands-on time of qPCR sample preparation
Andrew prepares qPCR samples with excellent accuracy and precision
After the full experimental setup done by Andrew or manually, the plates were assayed in an iCycler iQ™ Real-Time PCR detection system (Bio-Rad). Standard curves were generated with serial dilutions of a reference cDNA preparation from murine spleen mRNA. For each data point, the Ct was calculated and analyzed. As the intersection between an amplification curve and a threshold line, Ct is a relative measurement of the concentration of the target in a PCR reaction: the lower the Ct value, the greater the amount of target nucleic acid in the sample [3]. No signals were detected for the samples without cDNA template (negative controls), indicating that no cross contamination occurred during automated qPCR set-up by Andrew. The linear regression of the qPCR data of the 4-dilution series for each gene was plotted and the trend lines were added in order to calculate the R2 (coefficient of determination). R2 is a statistical measurement of how well the regression line approximates the real data points: the higher the R2 values, the better the accuracy. All qPCR data produced by Andrew and manually had the R2 values above 0.99, indicating supreme accuracy, with data from Andrew in general at higher values (Fig. 5). These results demonstrate that Andrew improves the pipetting accuracy of the serial dilutions compared to those obtained manually.
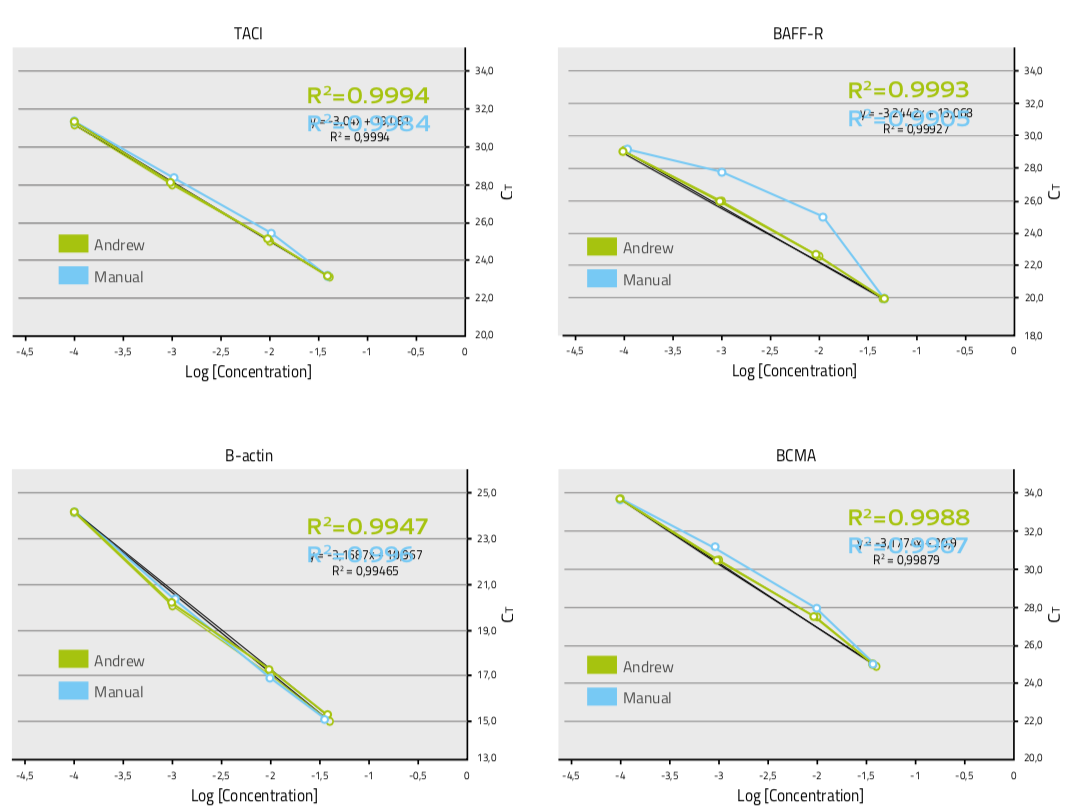
Figure 5: Pipetting accuracy measured as linear regressions of Ct from qPCR of the dilution series of 4 genes produced by Andrew and manually
The reproducibility of pipetting was also calculated as the coefficient of variations (CVs) of the Ct for duplicates and triplicates. The CVs of Ct obtained with Andrew ranged 0.11% – 0.17%, while the CVs of Ct obtained manually varied much greater, between 0.07% and 0.3%, suggesting that Andrew’s pipetting is more consistent than that of the experienced human operator (Fig. 6).
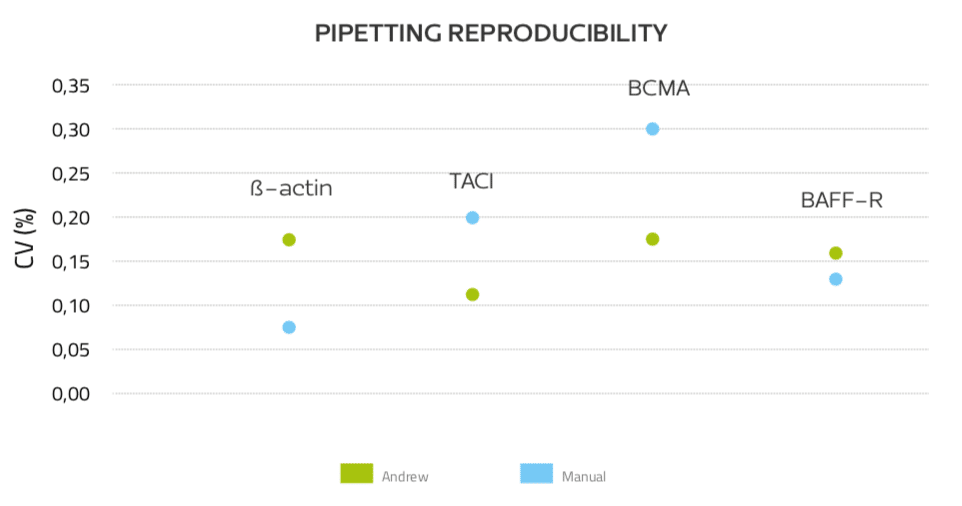
Figure 6: Pipetting reproducibility of Andrew and manually
Summary
Andrew stands as the most affordable and simple-to-use robotic platform for the set-up of qPCR experiments while maintaining the supreme accuracy and reproducibility that are required for this type of sensitive quantitative assays. The pipetting performance of Andrew allows scientists to standardize qPCR setup of their reactions free of errors and cross contamination with absolute ease. In addition, the required hands-on time necessary to prepare the experiment was significantly reduced (only 12 minutes for designing protocol and preparing consumables for Andrew to execute the pipetting). By utilizing Andrew for these processes, laboratories can now significantly improve both lab efficiency and quality of downstream results without any modifications to their existing workflows.
Acknowledgments
We thank Dr. ML Santiago-Raber (Faculty of Medicine, Depart- ment of Pathology and Immunology, University of Geneva, Switzerland) for the reagents and her expert technical assistance in preparing and analyzing the qPCR experiments.